Overview: mechanical signaling pathways regulating the structure & function of the heart
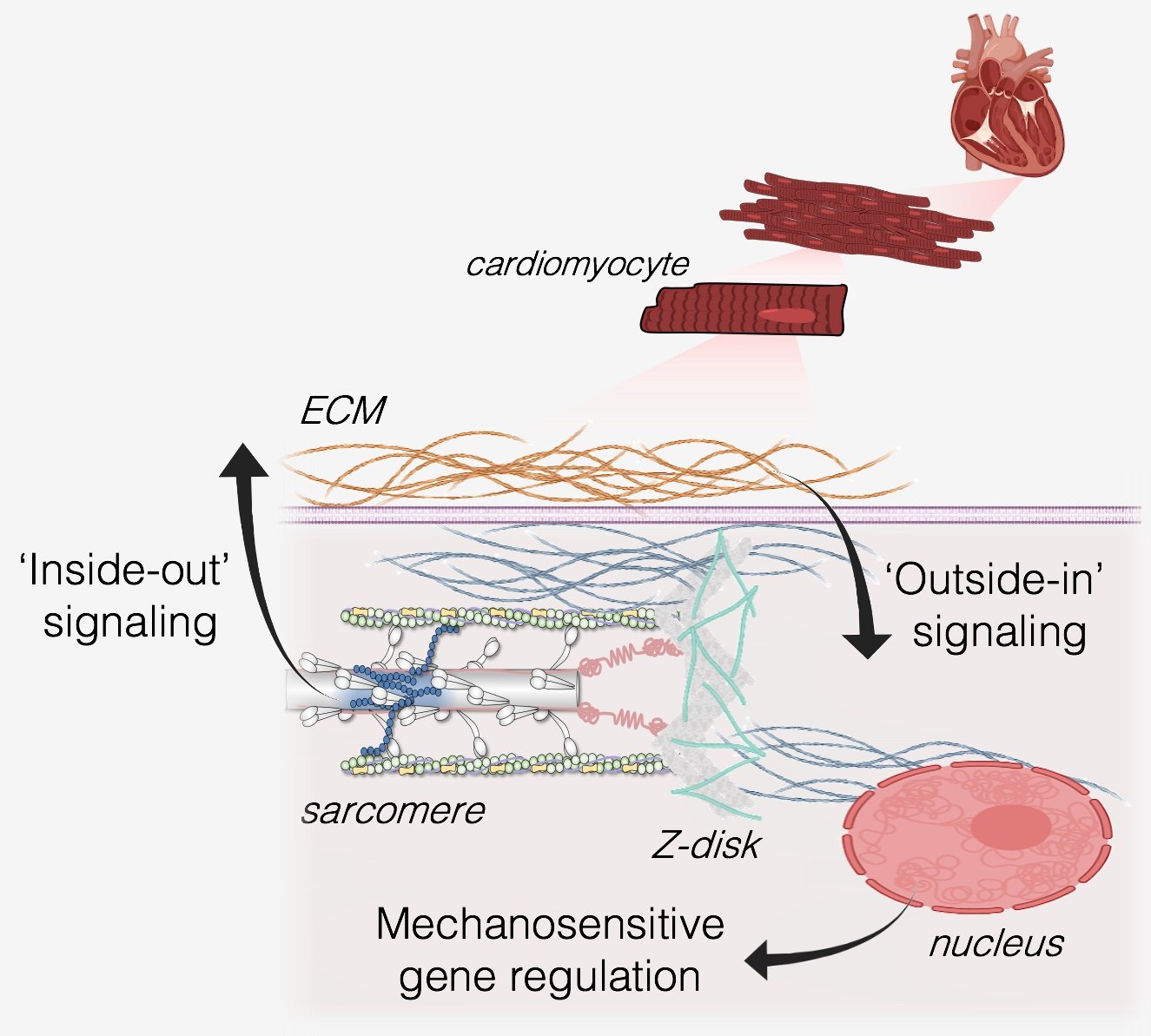
With every heartbeat, heart muscle cells (cardiomyocytes) are exposed to extreme mechanical forces and deformations. In heart disease, the magnitude and distribution of these forces can become abnormal, which often progresses the disease into heart failure. Our lab is focused on understanding the ways in which cardiomyocytes produce, sense, and process mechanical signals in healthy and diseased hearts, with the greater goal of discovering new therapeutic targets to prevent or reverse heart disease.
Contractile forces from inside the cell are generated primarily by molecular motors in the sarcomere, and these forces are transmitted throughout the cell and outward to the extracellular matrix (ECM) which we refer to as ‘inside-out’ mechanical signaling. Forces from outside the cell (from the extracellular matrix and/or passive stretches during ventricular filling) are transmitted into the cells during ‘outside-in’ mechanical signaling. In either case, mechanical forces pass between the extracellular matrix, the cytoskeleton, and the nucleus, where they can affect genetic programs that regulate the structure and function of the heart.
-
The big questions that motivate our research are:
What are the molecular signaling pathways involved in transmitting forces into and out of heart cells?
How are these mechanical signals ‘sensed’ by molecular and macromolecular machinery in heart cells and transduced into genetic programs that mediate cardiac growth/remodeling?
To what extent do defects in these mechanosignaling pathways, caused by mutations in the proteins that comprise them, drive the development of specific forms of heart diseases?
By better understanding the molecular/cellular mechanobiology of the heart, can we develop new therapies for heart diseases that target specific nodes in the mechanosignaling pathways of heart cells?
-
We take a multidisciplinary approach that encompasses elements of pathology, engineering, biophysics, and computational modeling, which enables us to collaborate with a broad scientific community from all over the world. We use genetically engineered rodent models and human stem cells to recapitulate aspects of various cardiac diseases in the lab.
-
Our main techniques include:
Experimental biomechanics on heart cells and tissue using both rodent models and human stem cell-derived cardiomyocytes.
Quantitative imaging methods such as X-ray diffraction techniques, electron microscopy, live-cell imaging, and immunofluorescent microscopy.
Multiscale computational models that help us interpret and analyze our experimental findings, predict physiological results, and generate new hypotheses for further investigation.
research foci
Multiscale biomechanics and biophysics of the heart
A common feature of most heart diseases is a change in the mechanical properties of the heart muscle. For example, certain heart diseases can alter the force-generating capacity of the ventricles during contraction or the passive stiffness during ventricular filling. Moreover, these changes at the ventricle level also manifest as changes in the cellular and molecular biomechanics. Discovering and understanding how disease-associated changes in cardiac mechanics span across such vastly different biological scales can help inform new therapies to prevent heart failure. As such, this area of our lab investigates the mechanical properties of the heart, from single proteins to the whole organ, and how these properties are affected in failing hearts. To do so, we use genetically engineered mouse models and human pluripotent stem cell-derived models of heart disease in a variety of experimental biomechanics assays. We quantify the contractile forces and passive mechanical properties of beating cardiac tissues and single cells with specialized equipment in the lab. We can combine these biomechanical and biophysical measurements with structural information and mechanistic computational models, which enables us identify new therapeutic strategies for heart diseases that reduce cardiac contractility, such as Dilated Cardiomyopathy (DCM).In this study, we targeted the integral of the twich tension-time relationship in DCM hearts using a combination of multiscale computational modeling and in-vitro experimental biomechanics. Through this approach, we prevented the development of DCM in mouse hearts (image from Powers et al., 2020).
Cellular mechanobiology underlying heart disease
Like many other cell types, heart muscle cells are very sensitive to the mechanical properties of their microenvironment. Extracellular mechanical signals are transmitted into the cardiomyocyte, where they are “sensed” and “processed” by various intracellular structures, including the nucleus, which transduce mechanical signals into biochemical and/or genetic outputs that regulate cell growth, structure, and function. Identifying these mechanosignaling pathways in cardiomyocytes, and how they fail in diseased hearts, is a major effort of our lab. To investigate this, we perturb the mechanics of the extracellular environment (for example, substrate stiffness, stretch, extracellular matrix composition) of gene-edited pluripotent stem cell-derived and/or rodent cardiomyocytes while monitoring intracellular remodeling. In parallel with these assays, we also use transcriptomic and proteomic analyses to characterize how extracellular mechanical stimuli and intracellular remodeling affect mechanically regulated gene expression in healthy and diseased cardiomyocytes. Through these studies, we investigate disease mechanisms in heart muscle cells that involve altered cell mechanosensing and mechanosignaling.Multiscale computational modeling of the heart in health & disease
A central philosophy of our research approach is that the combination of experimental studies and computational tools is critical for deepening our interpretation of new results, fueling hypothesis generation, predicting outcomes, and spanning orders-of-magnitude spatiotemporal scales. As such, we use and develop computational models of muscle biophysics that range from atomic scales to tissue-level scales. We can inform these models with experimental biomechanics data and multiscale imaging modalities (for example, X-ray diffraction techniques with live cells to measure nanometer-scale structural dynamics of sarcomere proteins, quantitative immunofluorescent imaging, electron microscopy) to recapitulate structural/functional properties of various muscle diseases in silico and predict potential restorative or preventative therapeutic targets.Human induced pluripotent stem cell-derived cardiomyocyte. Image courtesy of Abby Nagle from the Davis Lab.
Spatially explicit computational model of a half-sarcomere. This model can simulate cardiac muscle twitch contraction dynamics in health and disease, aiding our ability to understand and therapeutical target underlying mechanisms of heart failure.